In this post, I want to discuss what drives me forward and explain my strong belief in graphs, showcasing a few examples where this technology has truly made a difference. My goal is to demonstrate the power of graphs while highlighting the significant gap between the fields of biology and medicine. Often, solutions for medical problems can be discovered in the biological domain through data exploration. I hope this post inspires others to seek out solutions and perhaps find ways to make the world a bit brighter.
Note: This post is using, but not meant to demonstrate, GraphExplorer (the project I am working on). Therefore, completeness and showcasing everything I have implemented are not mandatory. While many parts of the implementation already exist, some details I will use to visualize my ideas are mocked and incomplete—they serve as a lighthouse for future development.
This specific story starts with a baby. At birth, everything was perfect, but by the time he was three months old, things began to go awry. Eventually, it was diagnosed that the causes were two different mutations in a gene called GFER. The exact medical details aren’t the focus here, so we can use analogies from computer science to simplify. Think of the gene as analogous to code, the resulting protein as the compiled code, and what the protein actually does as the compiled code deployed as a microservice.
The first mutation causes the destabilization of the protein, which is equivalent to our microservice experiencing runtime errors and crashing under pressure or over time. The second mutation is a premature stop codon. In our computer science analogy, this is like having a mistake in our code where an “AND” was replaced with an “END” inside a condition. A single letter change, but when the code tries to compile and reaches the “END,” the program stops running, resulting in partly processed results.
GFER, in itself, is a critical infrastructure “service,” and the entire system cannot function without it.
Back to the child: the medical world has encountered fewer than a dozen cases with problems in this gene, all with severe consequences. Facing the lack of any known way to help, the medical advice was limited to describing how things would continue to deteriorate, wrapped in phrases like “love the child while you can.”
Fast forward a few years, and the child is about to start first grade. He is intelligent, empathetic, strong, and healthy. While the future still holds its perils, uncertainty, and fear, it also carries lots of laughter, love, and hope.
This change of course happened through much hard work done mostly manually (and lots of luck). Then, a few years later, I encountered a technology with the potential to reduce thousands of research hours into dozens and bring hope to many more cases like this. I immediately fell in love with it, knowing it can help children and their families who suffer along with them. The ability to bring some light into these dark places truly matters.
How does this relate to data? In retrospect, everything was modeled as graph data, which allowed for continuous progress by placing each piece of information in its proper context until things started to “click together” and solutions emerged. This approach will be illustrated with examples of how such data exploration can be done. It is assumed that the data already exists and is “glued” together properly (data professionals are probably rolling their eyes with a “I wish it was so easy” expression).
Let’s dive into the data and specific examples. Remember the first problem? It was a stability issue with our protein (or microservice). Simplifying things, all we care about is the end result. In the case of proteins, just like microservices, there isn’t a single one doing all the work but many copies working together, sharing the load. Therefore, if we could consistently start more of these microservices, each individual microservice crash would be less of an issue as others could take its place. In biology, this can be achieved through transcription factors, which can be activated by external compounds in chains of causes and effects.
As I see it, in the ideal way of working (and in the system I am developing), finding these transcription factors should be achievable through a simple query. This query should be easily and visually modeled by anyone familiar with the subject (to ask what they want) and the data available to them (probably a bioinformatics or medical analyst). A query should be modeled by just stating what is known and what is sought; these will form a set of constraints over the data to be retrieved.
It will look similar to this: The query:
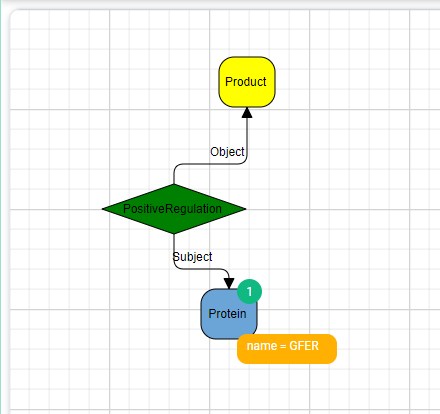
- State the Known Information: We have an entity of type Protein which has an attribute called name and this attribute has the value of “GFER”.
- Define the Desired Outcome: We need to find a Chemical Entity that positively regulates this protein. In the query above it is represented as constraining the answer to have an entity of type ChemicalEntity and that is it, in this query, we didn’t put any other bound on this entity.
- Set Constraints and Relationships: The protein (GFER) must be connected to a ChemicalEntity by a Positive Regulation relation and vice versa the ChemicalEntity must be bound to GFER through a positive regulation relation and by the role of “Object”.
That’s it, so easy to make.
The colors in the diagram are derived from visual customization set by the client, allowing them to assign (per data schema) a color to each type of entity and relationship.
While I didn’t run this specific query (yet), the result would look similar to the example shown in the “On Inference Rules Over Graphs” post. It will form a “star” like graph, with the protein (GFER) at the center and many Chemical Entities connected to it through Positive Regulation relationships.
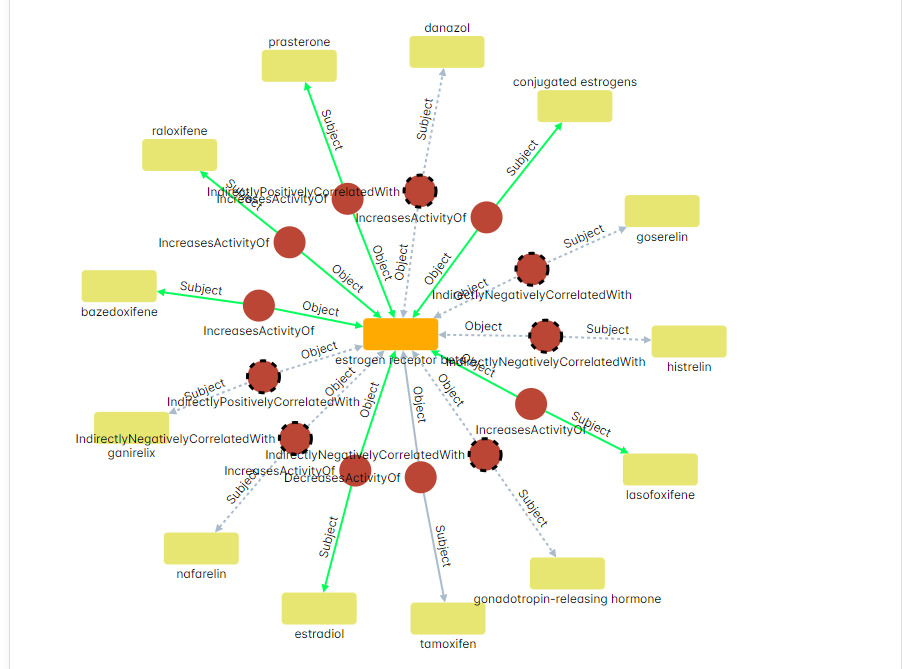 In this specific case, all of the connections will be inferred (represented by dashed lines) since the relation is mediated through the activation of a transcription factor named Nrf2. Nrf2 can be activated via multiple pathways (e.g., PI3K, ERK), leading to the inferred connections between the protein (GFER) and the various Chemical Entities. This inference is done implicitly in the background (though logic not statistical Machine Learning) and can be explained through a single click (for more information see the example in the “On Inference Rules Over Graphs” post).
In this specific case, all of the connections will be inferred (represented by dashed lines) since the relation is mediated through the activation of a transcription factor named Nrf2. Nrf2 can be activated via multiple pathways (e.g., PI3K, ERK), leading to the inferred connections between the protein (GFER) and the various Chemical Entities. This inference is done implicitly in the background (though logic not statistical Machine Learning) and can be explained through a single click (for more information see the example in the “On Inference Rules Over Graphs” post).
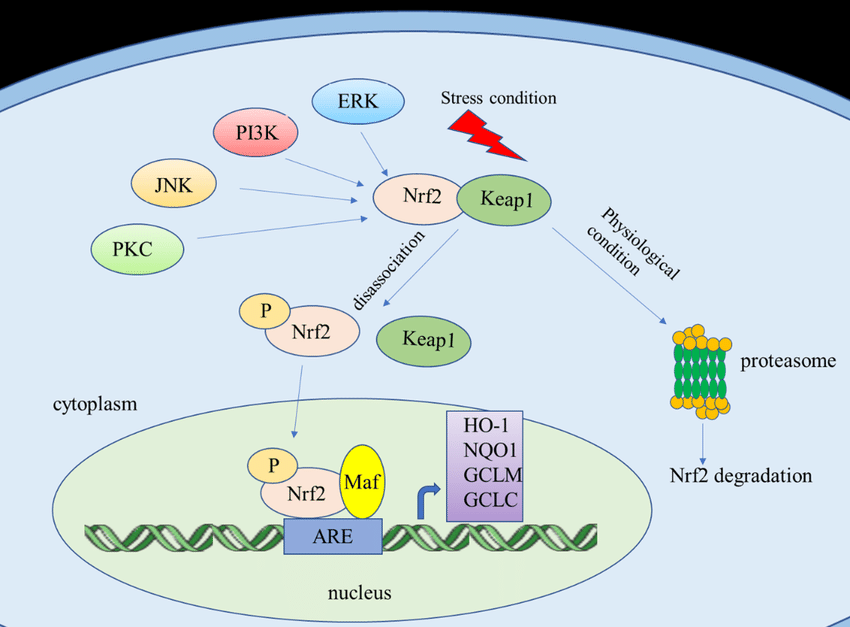 — Food-Derived Pharmacological Modulators of the Nrf2/ARE Pathway: Their Role in the Treatment of Diseases. DOI: 10.3390/molecules26041016
— Food-Derived Pharmacological Modulators of the Nrf2/ARE Pathway: Their Role in the Treatment of Diseases. DOI: 10.3390/molecules26041016
We can easily take it further. Let’s examine the following query:
If we have the data that a product contains the Chemical Entity, the query will work, though the explanation path becomes somewhat longer. This is because the query will need to trace the relationship from the product to the chemical entity and then to the protein through the inferred pathways. Again, all the inference is done implicitly in the background but can be fully explained.
Why does this matter?
In just a few clicks, it becomes possible to find potential new solutions. To help perceive the impact of this specific example, in reality, the activation of Nrf2 regulates more than 250 genes, with GFER being just one of them. There are likely many others who can reuse any solution related to this transcription factor.
This information is widely available from a biological perspective but is often missed in the medical domain, as the journey from research to medicine (“bench to bedside”) can take a decade or more. Please note that rushing this journey carries its risks, and it is important to seek professional consulting and maintain vigilant oversight before deciding to walk this path.
Let’s take another example. In the problem description before, we talked about the replacement of AND with END (biologists call this a premature stop codon, or PTC). What if we could replace our compiler with a “contextual error-correcting compiler” that detects the PTC is out of context and ignores the END command?
Well, in biology, this is somewhat possible through a process called PTC readthrough. Can we build a query that will help us find how to activate this process? Yes. Again, there are molecules that facilitate this, and there is a super simple query to find them.
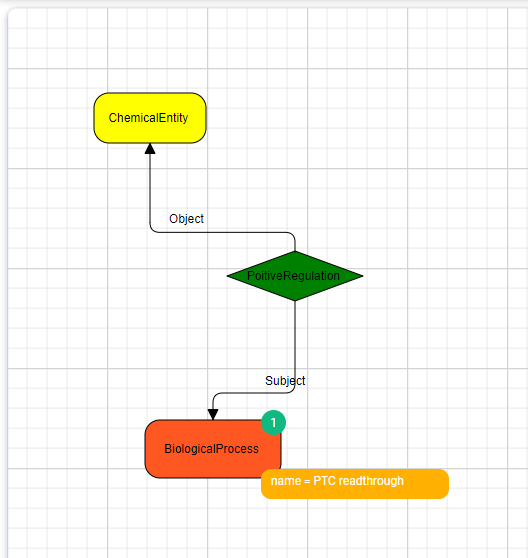
Here, reader, I need your help. I believe that this way of creating queries makes critical information easily approachable while keeping it explainable. It is visual and intuitive to learn, and with a predefined schema, the system’s assistance in building the queries can go a long way. The queries I presented were simple, but more advanced queries will be reviewed in future posts.
However, this is my belief. Your opinions, ideas, and challenges can significantly enhance this approach. If you have insights on making query building easier or more powerful, please share them with me. Your feedback can make a big difference.
When modeling all of this manually, additional abstraction layers were added alongside the biological ones (it is in the roadmap). The resulting abstraction layer looked like this small section:
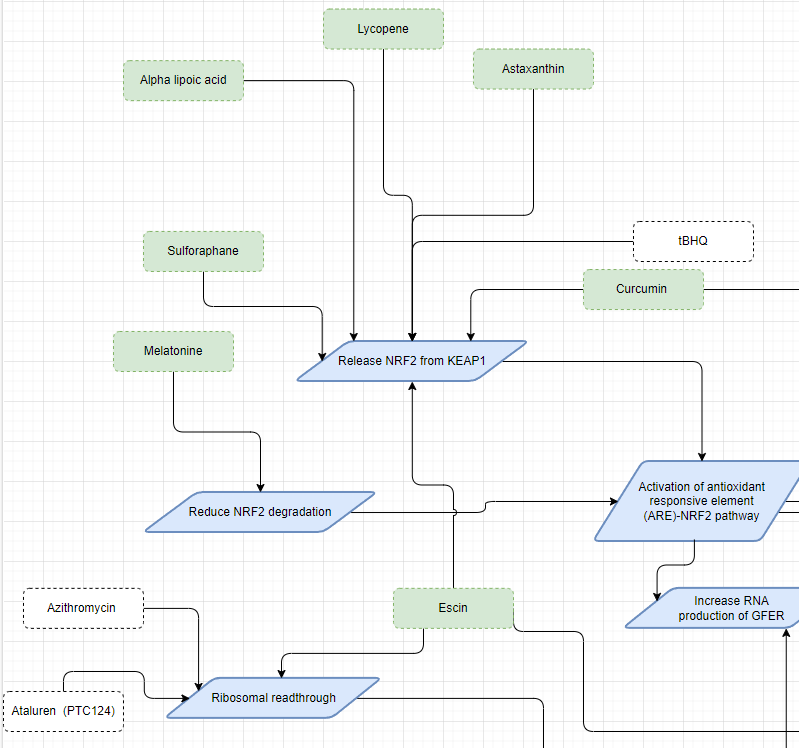
One interesting thing to notice is that when putting the results of the two solution strategies together, there exists a substance called Escin. Escin is connected to both the increased regulation of GFER production and the activation of PTC readthrough, making it a potentially powerful tool in our toolbox.
Suddenly, we have choices where before there were none. This example can be extended to improve GFER stability, reduce stresses on GFER, mitigate damages in the mitochondria, cells, and the whole body, and facilitate rapid repair of damages, among other possibilities. At every step, more options and tools become available. Working with these tools and searching for synergies has led to significant changes in a child’s life. For me, it provided an answer to why and how what I do matters.
Revealing these options took an endless amount of time, making it un-reproducible. But now it is possible to build solutions that will make such processes much more accessible. I hope this will help banish some darkness from the world, and for me, that matters!
I hope you found the post interesting from a data perspective. If you are facing a medical issue, I hope it gave you some hope and motivation to take action and explore further.
You are welcome to follow me as I continue to build TypeDB GraphExplorer, and feel free to contact me with any questions.
Currently, I am looking for ideas on how to make this development sustainable and for design partners (not necessarily in the bio/medical domains) to collaborate with and create clear value together.
